Workshop on Metadata for raw data from X-ray diffraction and other structural techniques
| Organized by | ||
| IUCr Diffraction Data Deposition Working Group | Croatian Association of Crystallographers | |
![[IUCr logo]](http://www.iucr.org/__data/assets/image/0007/61378/iucr.gif) |
![[Croatian Association of Crystallographers logo]](http://www.iucr.org/__data/assets/image/0014/104414/HUK_LogoDugiSrednji.jpg) |
|
Saturday August 22 - Sunday August 23 2015
Hotel Park, Arupinum Hall, Rovinj, Croatia
This two-day Workshop is organised by the DDD Working Group (WG), appointed by the IUCr Executive Committee to define the need for and practicalities of routine deposition of primary experimental data in X-ray diffraction and related experiments. It will take the form of a two-day satellite of the 29th European Crystallographic Meeting with lectures from crystallographic practitioners, data management specialists and standards maintainers.
Objective: As part of the continuing activities of the IUCr Diffraction Data Deposition Working Group, this workshop will seek to define the necessary metadata that needs to be captured and deposited alongside experimental diffraction images in order that such raw data may be subsequently re-evaluated or re-used in more detailed scientific studies. The workshop will also explore the metadata requirements of other structural experimental techniques used by crystallographers.
There is a public forum for discussion of the issues covered in this workshop at http://forums.iucr.org (and viewable in the 'Forum' tab below).
Acknowledgements: As the scope of this Workshop has broadened from its original conception, we are very grateful to various research institutes and universities and to CODATA for sending their staff to take part in this event. Support for technical services and associated staffing costs has been contributed by Dectris, IUC Journals, CODATA, the Cambridge Crystallographic Data Centre, Bruker, FIZ Karlsruhe/ICSD, Oxford Cryosystems and Wiley, to whom we are very grateful, as well as to the IUCr. We are also very grateful to the Croatian Association of Crystallographers for their active help in securing the best possible Workshop to address this important topic.
Programme
Saturday 22 August |
|||
| 10:00 | Open | ||
Session I: Introduction |
|||
| 10:00-10:05 | John R. Helliwell and Brian McMahon | Introduction and Welcome | |
| 10:05-10:20 | John R. Helliwell | Update on activities of the IUCr Diffraction Data Deposition Working Group (DDDWG) | Abstract | Presentation |
|
John R Helliwell1 and Brian McMahon2 This workshop follows on from the 2012 Workshop on Diffraction Data Deposition in Bergen (http://www.iucr.org/resources/data/dddwg/bergen-workshop) and Working Group meetings at ECM28 (U. Warwick, UK; see http://forums.iucr.org/viewtopic.php?t=332) and the IUCr Congress (Montreal, Canada; http://forums.iucr.org/viewtopic.php?t=347). The Bergen Workshop identified the need for a thorough examination of current practice with metadata for raw diffraction data, and the possibility of using such a review to stimulate improved metadata characterization and handling in non-diffraction studies. This workshop will address both requirements. In the wider scene 'Open Data' as a requirement of research publication is accelerating, whether it be derived, processed or raw data. Crystallography as a field compares well with other fields such as astronomy and particle physics in achieving 'open data' and each field finds raw data archiving challenging, especially the square kilometre array (SKA) in radio astronomy, since raw data is obviously the most voluminous. However, volume alone is not the greatest challenge. Stored raw data must be properly described so that its value and reliability can be assessed and understood, and individual data sets must be discoverable and reusable by other researchers, whether associated with formal publications or not. This is where metadata plays a key role. We addressed technical options for achieving raw data archiving in Bergen and favoured flexibility in the physical location of the data sets, but with a key need for assigning DOIs to each raw data set. Interestingly, Nature magazine on 9 July9th 2015 highlighted 'the cloud' and commercial providers as being a preferred method for genomics data archiving. The change of attitude of the USA NIH, for example, where there were worries over the security of the commercial data store option, is significant. (hide | hide all) |
|||
Session II: Diffraction images - what can we get out? |
|||
| 10:20-11:00 | Keynote: Loes Kroon-Batenburg | The need for metadata in archiving raw diffraction image data | Abstract | Presentation |
|
Loes M.J. Kroon-Batenburga & John R. Helliwellb Recently, the IUCr (International Union of Crystallography) initiated the formation of a Diffraction Data Deposition Working Group with the aim to develop standards for the representation of raw diffraction data associated with the publication of structural papers. Reports and minutes of DDDWG meetings can be found at forums.iucr.org. Archiving of raw data serves several goals: to improve the record of science, to verify the reproducibility and to allow detailed checks of scientific data, safeguarding against fraud or to allow reanalysis with future improved techniques. In a special series of papers on "Archiving raw crystallographic data" (Terwilliger, 2014), we reported on our experience of transferring and archiving raw diffraction data and on the problems encountered with acquiring and deciphering sufficient metadata (Kroon-Batenburg & Helliwell, 2014). To be able to process the raw data one needs information on the pixel geometry, information on pixel wise corrections applied, on beam polarization, wavelength and detector position amongst others, which are ideally contained in the image header. We will demonstrate that often one needs prior knowledge, evidently of how to read the (binary) detector format, but also on the set-up of goniometer geometries. This raises concerns with respect to long-term archiving of raw diffraction data. Care has to be taken that in the future unambiguous information is available i.e. one cannot simply "deposit the raw data" without such metadata details. We made available a local raw X-ray diffraction images data archive at the Utrecht University (rawdata.chem.uu.nl), subsequently mirrored at the Tardis Raw Diffraction Data Archive in Australia, and since March 2015 made available through digital object identifiers (doi) at the eScholar University of Manchester Library data archive. Since 2013 approximately 150 GB of data was retrieved from our archive and some of the data sets which were reprocessed by other groups. Kroon-Batenburg, L.M.J. & Helliwell, J.R. (2014) Acta Cryst. D70, 2502-2509. |
|||
| 11:00-11:20 | Coffee | ||
| 11:20-11:45 | Wladek Minor | Crystallographic raw data: our plans and implementations within the NIH's Big Data to Knowledge resource | Abstract | Presentation |
|
The NIH pilot project 'Integrated resource for reproducibility in macromolecular crystallography' will create a web-based archive of diffraction images collected from macromolecular samples around the world. The resource will enhance and sustain the macromolecular diffraction data comprising the primary data sources for macromolecular atomic coordinates in the Protein Data Bank (PDB). The project will develop tools that will extract metadata from images alone, or from a combination of information obtained from a PDB deposit and diffraction images. All of the metadata needed for automatic determination and re-determination of macromolecular structures will be collected. Currently, the project has more than 1500 data sets and a preliminary system for extracting certain types of metadata. The data mining tools developed will allow for analysis of single experiments, as well as sets of experiments performed using various synchrotron and home based sources. Diffraction sets and metadata will be available from the project's website at http://www.proteindiffraction.org, or through a link on a PDB deposits page on the RCSB PDB website. This talk will present initial results of data mining performed on the archive. (hide | hide all) |
|||
| 11:45-12:10 | Michael E. Wall | Metadata needed for the full exploitation of diffuse scattering data from protein crystals | Abstract | Presentation |
|
Michael E. Wall I will review efforts to model motions of crystalline proteins using diffuse X-ray scattering. This work requires analysis of raw diffraction images, which are mostly inaccessible in public databases. There is an abundance of potential metadata from these studies, including information about the analysis methods, measurements of diffuse intensity, and results from the modeling. The time is now ripe for integrating diffuse scattering into traditional crystallography: modern beam lines and detectors are enabling higher quality data collection; computations which were previously inaccessible are now becoming feasible; and current protein crystal structure determination methods are approaching the limit of what is possible using the Bragg peaks alone. The deposition of raw images and associated metadata in public databases is a key step in enabling analysis of diffuse scattering for all protein crystallography studies. (hide | hide all) |
|||
| 12:10-12:35 | Natalie Johnson and M. R. Probert | X-ray Origins: Protection or Paranoia? | Abstract | Presentation |
|
Natalie Johnson1 and Michael R. Probert1 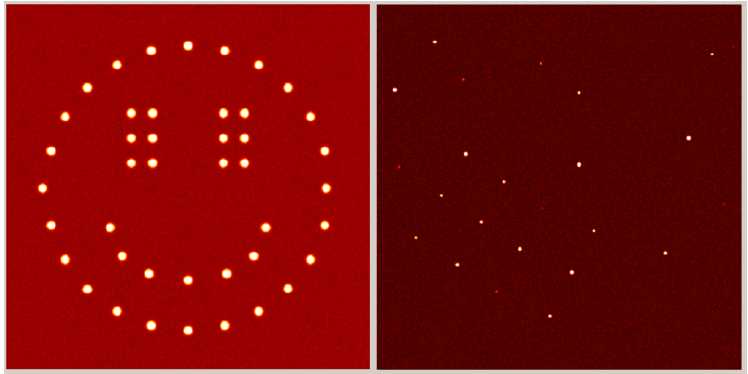 Figure 1. Two diffraction images - which is real? Deliberate fabrication of crystallographic data has previously led to falsified structures being published and then later retracted from respected scientific journals1-3. Identified perpetrators, in these cases, had made very simple modifications to structural files, such as manually changing unit cell sizes and atom types, to produce adjusted data. Fortunately they were found to be unable to produce raw experimental data to support their claims. Kroon-Batenburg and Helliwell4 proposed that the requirement for the deposition of raw crystallographic data may be a potential method of preventing the submission of counterfeit structures. However we can show that the recreation of raw diffraction images is no longer difficult, opening the doors for those less scrupulous to take advantage, if this is not already occurring! Detector frame formats from many manufacturers are well documented and this information can be reverse-engineered to encode synthetic diffraction data. This process was brought to light as a product of research into optimising data collection parameters for charge density studies. The chosen method required us to produce an algorithm which takes data from integrated .raw files as a starting point to create replicas of experimental images. A simple misuse of this code could take structure factors calculated for an entirely fabricated compound and produce diffraction images that, when processed, return the artificial structure. The frames are not visually distinguishable from authentic, experimentally determined, ones and can be fully integrated using standard protocols. The authors find this situation potentially alarming and requiring immediate attention. A structure refined from data processed from these artificial diffraction images could pass all IUCr checkCIF5 protocols without raising alerts. We will present such a structure, full details of the algorithms employed and propose methodologies that may safeguard against this approach going undetected. 1. T. Liu et al. (2010). Acta Cryst. E66, e13-e14. Keywords: Data, Simulation, Software, (hide | hide all) |
|||
| 12:35-13:00 | Andreas Förster | EIGER HDF5 data and NeXus format | Abstract | Presentation |
|
Andreas Förster and Marcus Müller, Dectris Ltd, Neuenhoferstrasse 107, 5400 Baden, Switzerland HDF5 is a container format designed for big data applications. In it, vast amounts of heterogeneous data can be stored in a small number of files that are easy to manage. Detectors of the EIGER series write datasets thousands of images big to HDF5 files and record most of the metadata that are required for data processing. The metadata are saved in a master file that is separate from the data but links to it. In this talk, I will present the HDF5 format and some of the metadata as written by EIGER detectors. I will also discuss metadata that are essential for processing but unknown to the detector and highlight blank fields that the EIGER HDF5 template provides for completion by beamline routines. A related talk by Herbert J. Bernstein [1] will explore ways of recording the geometry of the experimental setup. Software development, data processing, and effective archiving will all benefit from strict adherence to standards set by the NeXus committee. [1] H. J. Bernstein "Metadata for characterising diffraction images: imgCIF and NeXus" in Workshop on Metadata for raw data from X-ray diffraction and other structural techniques, 22 - 23 Aug 2015, Rovinj, Croatia. (hide | hide all) |
|||
| 13:00-14:00 | Lunch | ||
Session III: Metadata for diffraction images and other experimental methods |
|||
| 14:00-14:25 | Herbert J. Bernstein | Common diffraction image metadata specification in imgCIF, HDF5 and NeXus | Abstract | Presentation |
|
Herbert J. Bernstein The introduction of a new generation of fast pixel-array detectors, such as the Dectris Eiger and the Cornell-SLAC Pixel Array Detector (CSPAD), has required us to revisit and extend approaches we have used in the past to represent the data (the diffraction images) and the metadata (the information needed to reconstruct the experimental environment within which the data were collected) [1] [2]. For example, the axis descriptions from the imgCIF (image-supporting-CIF) dictionary have proven effective in reliably preserving the information about the frame-by-frame relative positions of beams, crystals and detectors and have been mapped into the context of HDF5 and NeXus to support the new Eiger format. See Andreas Förster's talk [3] for a discussion of the Dectris Eiger-specific HDF5/NeXus format. We are introducing a new, extended templating scheme to allow each beam line to specify the unique characteristics that will allow the metadata for a beam-line to be specified as either an HDF5/NeXus file or as an equivalent CBF/imgCIF file from which a site-file to be merged with run-specific data and metadata will be generated. A central repository of site-templates will be offered for convenience. This approach will help both in ensuring ease of processing of original data and in facilitating reliable handling of archived data. [1] H. J. Bernstein, J. M. Sloan, G. Winter, T. S. Richter, NIAC, COMCIFS, "Coping with BIG DATA image formats: integration of CBF, NeXus and HDF5", Computational Crystallography Newsletter, 2014, 5, 12-18. |
|||
| 14:25-14:50 | Andrew Götz | Towards a generalised approach for defining, organising and storing metadata from all experiments at the ESRF | Abstract | Presentation |
|
After more than 20 years of operation the situation concerning metadata at the ESRF is still very disparate between beamlines. The way the metadata required to analyse the raw data are defined and collected depends largely on the beamline concerned. Approaches vary from fully automated solutions implemented on the MX beamlines to a combination of automated and manual collection of metadata. This talk will not present the solution for MX (see talk by Gordon Leonard for more info) but will present a new approach for automating the collection and storing of well defined metadata for all experiments. The solution is based on a generic tool built at the ESRF which uses HDF5 for file format, Nexus for definitions (where possible) and icat for the metadata catalogue. The talk will present concrete examples of its use for nano tomography and fluorescence and radiation therapy. Ongoing work on how this will be extended to small angle scattering, coherent diffraction and eventually all other techniques will be presented. The talk will conclude with a discussion on the role of metadata in data policy and management. (hide | hide all) |
|||
| 14:50-15:15 | John Westbrook | The PDB and experimental data | Presentation |
| 15:15-15:35 | Tea | ||
| 15:35-16:00 | Tom Terwilliger | Realising the Living PDB and how raw diffraction data and its metadata can help | Abstract | Presentation |
|
Thomas C. Terwilliger1 and Gerard Bricogne2 The Protein Data Bank (PDB) is the definitive repository of macromolecular structural information. The availability of structure factors for most entries in the PDB has made it possible to continuously improve the models in the PDB by reinterpreting the primary data for existing structures as new methods of analysis, new biological information, and new ways of describing structures become available. This continuous improvement will be even more powerful once the diffraction images associated with each entry become accessible. The key factor is that depositing raw images will stimulate the improvement of integration and processing software in the same way as the deposition of merged X-ray data hugely stimulated progress in refinement software. Revisiting deposited images with that improved software will deliver more accurate data (especially, free from the currently inadequate treatment of contamination by multiple lattices) against which to re-refine the deposited structures themselves. With the initial interpretation of a structure, the original structure factors and the raw images, it will become possible both to carry out extensive validation of structures and to apply new algorithms for structure determination and analysis as they become available, leading to structures of ever-increasing accuracy and completeness. Keywords: Structure quality; validation; PDB; automation; structure determination; raw data deposition (hide | hide all) |
|||
| 16:00 | Close | ||
Sunday 23 August |
|||
| 09:00 | Open | ||
Session IV: Data in the Wider World - From Laboratory to Database |
|||
| 09:00-09:25 | Simon J. Coles | Diffraction Data in Context: metadata approaches | Abstract | Presentation |
|
Simon Coles Diffraction experiments and the results arising from them must often sit in a given scientific context - e.g. in chemical crystallography, they are often performed as part of a study concerned with synthesising and characterising new compounds. The context for an experiment, i.e. why it has been performed, is often lost - particularly in the case where data is published on its own. I will present approaches not only to ascribing metadata to the results of crystallographic experiments, but also to the general chemistry leading up to them. The first stages of work to build a model to support this have been published - http://www.jcheminf.com/content/5/1/52. I will go on to discuss recent work in two projects: (1) a collaboration with five big pharma companies, instrument manufacturers, electronic lab notebook vendors and the Royal Society of Chemistry to derive metadata for capturing the 'process' of performing experiments; and (2) a project (https://blog.soton.ac.uk/cream/) aimed at using metadata actively in the process of performing research, as opposed to purely for archival purposes. I will conclude with insights as to how the approaches taken in assigning metadata in these projects are important to consider when archiving and disseminating raw crystallographic data. (hide | hide all) |
|||
| 09:25-09:50 | Suzanna Ward | CCDC metadata initiatives | Abstract | Presentation |
|
Suzanna Ward, Ian J. Bruno, Colin R. Groom and Matthew Lightfoot For half a century the Cambridge Crystallographic Data Centre (CCDC) has produced the Cambridge Structural Database (CSD) to allow scientists worldwide to share, search and reuse small molecule crystal structure data. An entry in the CSD is often seen as 'just' a set of coordinates, but the associated metadata (data that describes and gives information about other data), is essential to contextualise an entry. Data that describes the substance studied, the experiment performed and the dataset as a whole are all vital. This presentation, timed to coincide with the 50th anniversary of the CSD, will look at how metadata is used from deposition to dissemination of the CSD. We will look at how recent developments surrounding metadata have been targeted to improve the discoverability, validation and reuse of crystal structure data before looking to see what the future may hold. (hide | hide all) |
|||
| 09:50-10:15 | Brian Matthews | Supporting Data Management Workflows at STFC | Abstract | Presentation |
|
Brian Matthews, Scientific Computing, Rutherford Appleton Laboratory, Science and Technology Facilities Council, Harwell Oxford, Didcot OX11 0QX, UK STFC has developed a systematic approach for managing and archiving data generated from its large-scale analytic facilities, which is used with variations by the ISIS Neutron Source, the Diamond Light Source and the Central Laser Facility. This is centred around the ICAT experiment metadata catalogue. The ICAT acts as a core middleware component recording and guiding the storage of raw data and subsequent access and reuse of the data; it has evolved into a suite of tools which can be used to build data management infrastructure. In this talk, I shall describe the current status of the ICAT. The data rates and volumes generated from facilities are ever increasing and experimental science is becoming more complex. This is presenting challenges to the user community in accessing, handling and processing data. I shall describe some approaches to these problems and consider how we are exploring further support for data analysis and publication workflows within a large-scale facility. Finally, I shall consider how we might develop metadata to capture and share this information across communities. (hide | hide all) |
|||
| 10:15-10:40 | Pierre Aller | Overview of metadata and raw data cataloguing at Diamond | Abstract | Presentation |
|
Pierre Aller and Alun Ashton, Diamond Light Source, Division of Science, Didcot, Oxfordshire, UK Diamond Light Source as a relatively new facility has been able to capture and catalogue all its raw data (now over 3.6 Petabytes). Additionally as much metadata and processed data as possible has always been collated with the raw data and captured into both databases (ISPyB) for query and quick access, and into the raw data files (imgCIF/CBF and NeXus). Progress and status on these developments will be presented. (hide | hide all) |
|||
| 10:40-11:05 | Brian McMahon | CODATA and (meta)data characterisation in the wider world | Abstract | Presentation |
|
Brian McMahon This Workshop concentrates on scientific metadata and their importance in maximising the utility, trustworthiness and reuse of scientific data, especially to open doors to further avenues of study, and even new scientific insight. In a more general context, 'metadata' is a vehicle for categorising, classifying and collecting data sets. This presentation will review some of the organisations that take an interest in generic metadata and interoperability between metadata specifications from different disciplines or communities. The CODATA/VAMAS Working Group on Description of Nanomaterials provides a good example of collating different specialist metadata elements in a broad interdisciplinary framework. There will be a brief discussion of the granularity mismatch between generic and specialised metadata systems. (hide | hide all) |
|||
| 11:05-11:25 | Coffee | ||
Session V: What new metadata items are needed? |
|||
| 11:25-11:50 | Gordon Leonard | What metadata is needed to make ESRF raw MX diffraction data intelligible for new users? | Abstract | Presentation |
|
Gordon Leonard The volume of diffraction data that can be collected during an experimental session on modern synchrotron-based Macromolecular Crystallography (MX) beamlines equipped with fast readout photon-counting pixel detectors and the rate at which it can be collected means it is currently difficult (or impossible) for users to manually process, during the experiment, all data sets collected. To help remedy this situation and to provide the at-beamline feedback that is sometimes necessary for a successful experiment 'autoprocessing' software[1,2] is often deployed with the results of automatic integration, scaling merging and reduction for individual data sets displayed in Laboratory Information Management Systems (LIMS) such as ISPyB[3] from where they can also be downloaded. While the 'autoprocessing' approach works (i.e. provides data of sufficient quality for structure solution and refinement) in the vast majority of cases there is an increasing need for the post-experiment processing of raw diffraction images. In such cases the correct metadata for each dataset are essential to ensure the best results. For MX diffraction data collected at the ESRF this is stored in ISPyB, in the headers of the raw data images themselves and in automatically generated input files for the two main packages - XDS and MOSFLM - routinely used in the processing of ESRF-collected MX diffraction data. During my talk I will review the metadata currently logged during MX experiments at ESRF and look forward to what further metadata might be required when, either for validation purposes or the testing of new data processing and analysis protocols, raw data images are routinely made available to the wider scientific community. [1] G. Winter et al. (2013). Acta Cryst. D69, 1260-1273. doi: 10.1107/S0907444913015308 |
|||
| 11:50-12:15 | Matthew Blakeley | What metadata is needed to make Institut Laue Langevin neutron diffraction raw data intelligible for new users? | Abstract | Presentation |
|
Matthew Blakeley Central facilities for neutron scattering and synchrotron X-ray sources in Europe are working together to develop and share infrastructure for the data collected there. Such co-operation should make it easier and more efficient for users to access and process their data, and provide more secure means of storage and retrieval. It should also increase the scientific value of the data by opening it up to a wider community for further analysis and fostering new collaborations between scientific groups. However, with these developments comes a need to define how raw data are stored and made accessible, and in particular, what metadata are included to allow diffraction data to be intelligible to new users. To this end, an ILL data policy (https://www.ill.eu/fr/users/ill-data-policy/) was established in 2012, and a number of tools (e.g. [i] https://data.ill.eu [ii] https://logs.ill.eu) are being developed. These currently allow experimental data (identified by a DOI) to be consulted and downloaded remotely and ultimately will allow for (re)processing and validation of experimental data. (hide | hide all) |
|||
| 12:15-12:40 | Kamil Dziubek | Metadata in high-pressure crystallography | Abstract | Presentation |
|
Kamil F. Dziubeka and Andrzej Katrusiakb The deposition of metadata related to specific techniques used for crystallographic experiments can be simplified by formulating guidelines for their preparation. One of the experimental techniques quickly gaining ground in the field of crystallographic research is high-pressure diffraction studies. They involve additional equipment for pressure generation, pressure calibration, etc. The high pressure cell can interfere with the primary or diffracted beam, which can contaminate the diffraction patterns and introduce errors in reflection intensities. The experimental details are vital for the evaluation and analysis of the data, and therefore the metadata are needed to be stored along with the raw diffraction images. The most essential descriptors concern: (1) orientation of the pressure cell with respect to the incident beam and the detector; (2) the sample preparation and shape, both for powder and single crystals; (3) a reference to the high-pressure vessel, and for unique equipment the dimensions of its relevant components, such as the anvil design, gasket thickness, chamber diameter, backing-plate type; (4) chemical composition of the cell parts, e.g. anvils, gasket and backing plates, pressure-transmitting medium; (5) the method of fixing the sample in the high-pressure chamber, if used; (6) the method of positioning the pressure cell during the data acquisition; (7) the pressure-measurement method. This information is indispensable for reproducing the results of structural refinements from the raw data or for attempting other methods of refinement. The pressure transmitting medium can dramatically change the sample compression, due to its possible interaction with the sample (such as penetration into the pores) or hydrostatic limit of the medium. The sample history can also affect the results. If the sample was recrystallized in situ in isothermal or isochoric conditions, from solution or melt, the details of the crystallization protocol should be provided. Simple edition rules and a checklist can considerably simplify the deposition of metadata and increase their informative value. The authors are representing the IUCr Commission on High Pressure, AK is the chair of the Commission. KFD gratefully acknowledges the Polish Ministry of Science and Higher Education for financial support through the "Mobilność Plus" program. (hide | hide all) |
|||
| 12:40-13:40 | Lunch | ||
Session VI: Metadata schemas |
|||
| 13:40-14:05 | James Hester | Creating and manipulating universal metadata definitions | Abstract | Presentation |
|
Metadata discussions are often closely linked to particular formats. However, facts about the natural world cannot depend on the medium used to transmit those facts. It follows that we are able to completely describe our metadata without reliance on any particular file format, and that we can distil metadata definitions from pre-existing data transfer frameworks regardless of the particular format used. This promising, if obvious, general conclusion does not specify what information needs to be provided in our format-free metadata definition. Following Spivak and Kent (2012) I suggest that it is sufficient that the metadata definitions can be expressed as functions mapping some domain to some range. This talk will explore some of the implications of this approach, including the independence of file format and metadata specification, specification of algorithms for interconversion between data files in differing formats, unification of disparate metadata projects, and simple steps to produce a complete metadata description. Spivak D.I., Kent R.E. (2012) "Ologs: A Categorical Framework for Knowledge Representation." PLoS ONE, 7(1):e24274. doi:10.1371/journal.pone.0024274 (hide | hide all) |
|||
| 14:05-14:30 | Brian McMahon | The Crystallographic Information Framework as a metadata library | Abstract | Presentation |
|
Brian McMahon The Crystallographic Information File (CIF) was introduced as a data exchange standard in crystallography in 1991 [1] and has become embedded in the practice of single-crystal and powder diffractometry. Versions of CIF exist that describe macromolecular structure and diffraction images [2], so that CIF may be used anywhere in a data pipeline from image capture to publication in what has been called a 'coherent information flow' (also CIF!) [3]. In practice, slight differences in data model and file format have led to 'dialects' of CIF, which can coexist quite happily. However, there is a consequent barrier to full interoperability. A new version of the CIF format [3] will allow the development of a new generation of CIF 'dictionaries' (the formal data description schemas or 'ontologies'). This will allow fully automatic interconversion between existing CIF data files in either formalism, but has the added bonus of providing a descriptive framework for any type of crystallographic information. Formally, the CIF approach makes no distinction between 'data' and 'metadata', and so is arbitrarily adaptable and extensible to any domain of structural science or further afield. Since the CIF format has a very simple syntactic structure which makes the contents very easy to read, CIF dictionaries can provide a simple template for developing new metadata schemas by working scientists who are not experts in informatics. [1] Hall, S. R., Allen, F. H. & Brown, I. D. (1991). The Crystallographic Information File (CIF): a New Standard Archive File for Crystallography. Acta Cryst. A47, 655-685. |
|||
| 14:30-14:40 | General discussion | ||
| 14:40-15:00 | Tea | ||
| 15:00-15:50 | Practical session: building a metadata description | Presentation 1 |
|
| 16:00 | Close | ||
| 18:00 | ECM29 Opening Ceremony | ||
 |
I. IntroductionBrian McMahon: Introduction and welcome ![[Brian McMahon]](https://www.iucr.org/__data/assets/image/0015/114108/mcmahon_1.jpg)
(05 min 06 sec) John Helliwell: Update on activities of the IUCr Diffraction Data Deposition Working Group (DDDWG) ![[John Helliwell]](https://www.iucr.org/__data/assets/image/0004/114169/helliwell.jpg)
(29 min 52 sec) II. Diffraction images - what can we get out?Loes Kroon-Batenburg: The need for metadata in archiving raw diffraction image data ![[Loes Kroon-Batenburg]](https://www.iucr.org/__data/assets/image/0014/114170/kroon-batenburg.jpg)
(39 min 11 sec) Wladek Minor: Crystallographic raw data: our plans and implementations within the NIH's Big Data to Knowledge resource ![[Wladek Minor]](https://www.iucr.org/__data/assets/image/0013/114160/minor.jpg)
(38 min 28 sec) Michael Wall: Metadata needed for the full exploitation of diffuse scattering data from protein crystals ![[Mike Wall]](https://www.iucr.org/__data/assets/image/0019/114157/wall.jpg)
(28 min 22 sec) Natalie Johnson: X-ray Origins: Protection or Paranoia? ![[Natalie Johnson]](https://www.iucr.org/__data/assets/image/0015/114162/johnson.jpg)
(21 min 46 sec) Andreas Förster: EIGER HDF5 data and NeXus format ![[Andreas Foerster]](https://www.iucr.org/__data/assets/image/0020/114158/foerster.jpg)
(30 min 22 sec) III. Metadata for diffraction images and other experimental methodsHerbert Bernstein: Common diffraction image metadata specification in imgCIF, HDF5 and NeXus ![[Herbert Bernstein]](https://www.iucr.org/__data/assets/image/0018/114156/bernstein.jpg)
(32 min 25 sec) Andrew Götz: Towards a generalised approach for defining, organising and storing metadata from all experiments at the ESRF ![[Andy Goetz]](https://www.iucr.org/__data/assets/image/0014/114161/goetz.jpg)
(28 min 59 sec) John Westbrook: The PDB and experimental data ![[John Westbrook]](https://www.iucr.org/__data/assets/image/0003/80337/jw.jpg)
(28 min 36 sec) Tom Terwilliger: Realising the Living PDB and how raw diffraction data and its metadata can help ![[Tom Terwilliger]](https://www.iucr.org/__data/assets/image/0003/114159/terwilliger.jpg)
(41 min 38 sec) IV. Data in the Wider World - From Laboratory to DatabaseSimon Coles: Diffraction Data in Context: metadata approaches ![[Simon Coles]](https://www.iucr.org/__data/assets/image/0010/114103/coles_480_360.jpg)
(33 min 42 sec) Suzanna Ward: CCDC metadata initiatives ![[Suzanna Ward]](https://www.iucr.org/__data/assets/image/0009/114111/WARD.jpg)
(25 min 17 sec) Brian Matthews: Supporting Data Management Workflows at STFC ![[Brian Matthews]](https://www.iucr.org/__data/assets/image/0013/114106/matthews.jpg)
(30 min 03 sec) Pierre Aller: Overview of metadata and raw data cataloguing at Diamond ![[Pierre Aller]](https://www.iucr.org/__data/assets/image/0012/114105/aller.jpg)
(18 min 59 sec) Brian McMahon: CODATA and (meta)data characterisation in the wider world ![[Brian McMahon]](https://www.iucr.org/__data/assets/image/0015/114108/mcmahon_1.jpg)
(33 min 56 sec) V. What new metadata items are needed?Gordon Leonard: What metadata is needed to make ESRF raw MX diffraction data intelligible for new users? ![[Gordon Leonard]](https://www.iucr.org/__data/assets/image/0014/114107/leomard.jpg)
(26 min 47 sec) Matthew Blakeley: What metadata is needed to make Institut Laue Langevin neutron diffraction raw data intelligible for new users? ![[Matthew Blakeley]](https://www.iucr.org/__data/assets/image/0008/114110/blakeley.jpg)
(24 min 23 sec) Kamil Dziubek: Metadata in high-pressure crystallography ![[Kamil Dziubek]](https://www.iucr.org/__data/assets/image/0011/114113/dziubek.jpg)
(23 min 44 sec) VI. Metadata schemasJames Hester: Creating and manipulating universal metadata definitions ![[James Hester]](https://www.iucr.org/__data/assets/image/0016/114109/hester.jpg)
(48 min 58 sec) Brian McMahon: The Crystallographic Information Framework as a metadata library ![[Brian McMahon]](https://www.iucr.org/__data/assets/image/0010/114112/mcmahon_2.jpg)
(22 min 56 sec) General discussion ![[banner]](https://www.iucr.org/__data/assets/image/0014/114116/dddwg_small.jpg)
(11 min 15 sec) |
|
Click on a thumbnail in the right-hand column to see the recorded presentation.
|
Workshop report
'Thank you for organizing such an inspiring workshop on what was nominally a rather dry subject.' Andreas Förster, Dectris
Indeed, the term 'metadata' – often described as 'data about data' or 'information to help you understand the data' - is generally held to be a dry topic, of importance to digital librarians and data analysts, but irrelevant or even an obstacle to the real business of science. This two-day satellite workshop of the 2015 European Crystallography Meeting demonstrated emphatically that this is far from the truth. Some 20 expert speakers from Europe, Australia and the USA (two presenting remotely over the Internet) surveyed the central importance of detailed and high-quality metadata to the interpretation, validation and use of experimental data.
![[workshop participants]](https://www.iucr.org/__data/assets/image/0016/114811/dddwg_workshop1.jpg) |
| Workshop participants enjoying a coffee break. |
The workshop was organised by the IUCr Diffraction Data Deposition Working Group (DDDWG) in association with the Croatian Association of Crystallographers. John Helliwell, Chair of the DDDWG, explained how it had been working over the past four years to analyse the prospects for routine deposition of raw experimental data, and had become aware that storage capacity for the vast amounts of raw data being generated at modern synchrotron and neutron facilities is almost the least of our worries. For this data to be re-used, it is essential that all details of the experimental arrangement are documented and retrievable – this is where 'metadata' comes into play.
Loes Kroon-Batenburg and Wladek Minor, among others, highlighted the very low level of standardisation in storing basic information about orientation, exposure, oscillation axis, etc. in the header of each image. The workshop renewed calls for agreement on a minimum set of such metadata that should be recorded in every image. Herbert Bernstein and Andreas Förster illustrated how the necessary definitions already existed in the imgCIF dictionary, and could effectively be carried over to the HDF5/NeXus files that are becoming the norm in high-volume imaging.
Current and evolving practice in data capture and management was described across a range of large-scale facilities accommodating a variety of techniques and sciences: the European Synchrotron, ESRF (Andy Götz, Gordon Leonard); Institut Laue–Langevin (Matthew Blakeley), the UK STFC and Diamond Light Source at the Rutherford Laboratories (Brian Matthews and Pierre Aller). Simon Coles spoke about the challenges of data management in home laboratories and medium-scale service providers such as the UK National Crystallography Service. In all these locations, all the data from an experiment must be handled in the context of resource management, provenance, validation and bulk storage, all of which require ever greater volumes of metadata that should conform to widely-accepted standards.
The importance to databases of carrying extensive metadata throughout the scientific process was described by Suzanna Ward (CCDC) and John Westbrook (PDB), while Tom Terwilliger developed the theme of 'The Living PDB', where deposited structures could be revised, improved and continuously updated in the light of new scientific developments. Mike Wall emphasised that exciting new science potentially lay in the diffuse scattering in images that is largely ignored when deriving structures solely from the Bragg peaks. Kamil Dziubek outlined the additional metadata that were needed to perform a complete analysis of structures collected under high pressure and other non-ambient conditions.
![[extracts from presentation]](https://www.iucr.org/__data/assets/image/0017/114812/dddwg_workshop2.jpg) |
| Montage of slides from Kamil Dziubek’s presentation (illustrations courtesy Ronald Miletich-Pawliczek, U. Vienna). |
In an intriguing presentation, Natalie Johnson demonstrated that plausible diffraction images could be manufactured. In principle, such artificial images could be produced to support fraudulent experimental results. Here, again, rich metadata describing the full provenance of the images and the context in which they were collected could help in forensic analysis of suspect data. Indeed, quite apart from worries about fraud, the more metadata that are available for cross-comparison, the more the data can be analysed (or reanalysed) for consistency, and the more trust can be placed in the scientific deductions that use the data.
The same considerations had encouraged the development by the IUCr of of checkCIF as a validation tool in the publication of crystal and molecular structures. There was a strong feeling in this Workshop that the time was rapidly approaching for the crystallographic community to work on a similar 'checkCIF' mechanism for the validation and evaluation of experimental data – perhaps a topic for the next DDDWG Workshop?
Perhaps most noteworthy is that the work of the DDDWG has become so much more urgent as raw data sets become increasingly available in the scientific environment. When this Workshop was first planned, rather few images were being stored on publicly-accessible platforms. Now, one may find raw data sets in repositories such as Australia’s Store.Synchrotron, on the NIH BD2K website http://www.proteindiffraction.org/ run by Wladek Minor’s group, on the shared resource site Zenodo, and in the powder pattern database maintained by the International Centre for Diffraction Data. Whether this growth will turn into a deluge of diffraction data sets is still unclear; what is certain is that the best use of such data sets will depend on metadata developments such as those explored during those two sunny days in Rovinj.
Brian McMahon
John R. Helliwell
About our sponsors
We acknowledge the generous financial support of the following partners.
 | Dectris is the technology leader in X-ray detection. The DECTRIS photon counting detectors have transformed basic research at synchrotron light sources, as well as in the laboratory and with industrial X-ray applications. DECTRIS aims to continuously improve measurement quality, thereby enabling new scientific findings. This pioneering technology is the basis of a broad range of products, all scaled to meet the needs of various applications. DECTRIS also provides solutions for customer developments in scientific and industrial X-ray detection. DECTRIS was awarded the 2010 Swiss Economic Award in the High-Tech Biotech category, the most prestigious prize for start-up companies in Switzerland. |
 | The International Union of Crystallography (IUCr) is a scientific union whose objectives are to promote international cooperation in crystallography and to contribute to the advancement of crystallography in all its aspects. The IUCr fulfills part of these objectives by publishing high-quality crystallographic research through nine primary scientific journals: Acta Crystallographica Section A: Foundations and Advances; Acta Crystallographica Section B: Structural Science, Crystal Engineering and Materials; Acta Crystallographica Section C: Structural Chemistry; Acta Crystallographica Section D: Biological Crystallography; Acta Crystallographica Section E: Crystallographic Communications; Acta Crystallographica Section F: Structural Biology Communications; Journal of Applied Crystallography; Journal of Synchrotron Radiation; and, launched for the International Year of Crystallography, IUCrJ, a gold open-access title publishing articles in all of the sciences and technologies supported by the IUCr. |
 | CODATA, the Committee on Data for Science and Technology, is an interdisciplinary Scientific Committee of the International Council for Science (ICSU), established in 1966. Its mission is to strengthen international science for the benefit of society by promoting improved scientific and technical data management and use. CODATA works to improve the quality, reliability, management and accessibility of data of importance to all fields of science and technology. CODATA provides scientists and engineers with access to international data activities for increased awareness, direct cooperation and new knowledge. It is concerned with all types of data resulting from experimental measurements, observations and calculations in every field of science and technology, including the physical sciences, biology, geology, astronomy, engineering, environmental science, ecology and others. Particular emphasis is given to data management problems common to different disciplines and to data used outside the field in which they were generated. |
 | Bruker Corporation has been driven by the idea to always provide the best technological solution for each analytical task for more than 50 years. Today, worldwide more than 6,000 employees are working on this permanent challenge at over 90 locations on all continents. Bruker systems cover a broad spectrum of applications in all fields of research and development and are used in all industrial production processes for the purpose of ensuring quality and process reliability. Bruker continues to build upon its extensive range of products and solutions, its broad base of installed systems and a strong reputation among its customers. Being one of the world's leading analytical instrumentation companies, Bruker is strongly committed to further fully meet its customers' needs as well as to continue to develop state-of-the-art technologies and innovative solutions for today's analytical questions. |
 | The Cambridge Crystallographic Data Centre (CCDC) is dedicated to the advancement of chemistry and crystallography for the public benefit through providing high quality information, software and services. Chemists in academic institutions and commercial operations around the world rely on the CCDC to deliver the most comprehensive and rigorous molecular structure information and powerful insights into their research. The CCDC is a non-profit organisation and a registered charity, supported entirely by software subscriptions from its many users. The CCDC compiles and distributes the Cambridge Structural Database (CSD), the world's repository of experimentally determined organic and metal-organic crystal structures. It also develops knowledge bases and applications which enable users quickly and efficiently to derive huge value from this unique resource. |
 | FIZ Karlsruhe is a leading international provider of scientific information and services. Our mission is to supply scientists and companies with professional research and patent information as well as to develop innovative information services. As a key player in the information infrastructure we pursue our own research program and also cooperate with leading universities and research associations. The Inorganic Crystal Structure Database (ICSD) is the world’s biggest database of fully evaluated and published crystal structure data. Science and industry are offered high-quality records that will provide a basis for studies in materials science, e.g. for identifying unknown substances. ICSD contains more than 165,000 crystal structures of inorganic substances published since 1913. Metals were included in ICSD several years. The metal structures were recorded retroactively in cooperation with FIZ Karlsruhe’s partner, NIST (National Institute for Science and Technology, Washington, DC, USA). FIZ Karlsruhe is a non-profit corporation and the largest non-university institution for information infrastructure in Germany. FIZ Karlsruhe is a member of the Leibniz Association, which comprises almost 90 institutions involved in research activities and/or the development of scientific infrastructure. |
 | Oxford Cryosystems is a market-leading manufacturer of specialist scientific instrumentation and software. The origins of the company lie in the design and manufacture of the original Cryostream Cooler in 1985, which immediately became the system of choice for cooling samples in X-ray diffraction experiments. The range of products for use in sample cooling has expanded over the last twenty-five years to include liquid-free systems, helium coolers and specially adapted systems for use with powder samples. Today the company is considered to be the global market leader in X-ray diffraction sample cooling. |
 | Wiley's Scientific, Technical, Medical, and Scholarly (STMS) business, also known as Wiley-Blackwell, serves the world's research and scholarly communities, and is the largest publisher for professional and scholarly societies. Wiley-Blackwell's programs encompass journals, books, major reference works, databases, and laboratory manuals, offered in print and electronically. Through Wiley Online Library, we provide online access to a broad range of STMS content: over 4 million articles from 1,500 journals, 9,000+ books, and many reference works and databases. Access to abstracts and searching is free, full content is accessible through licensing agreements, and large portions of the content are provided free or at nominal cost to nations in the developing world through partnerships with organizations such as HINARI, AGORA, and OARE. |


![[J. R. Helliwell]](https://www.iucr.org/__data/assets/image/0003/113736/jrh.jpg)
![[L. Kroon-Batenburg]](https://www.iucr.org/__data/assets/image/0014/113351/loes.jpg)
![[W. Minor]](https://www.iucr.org/__data/assets/image/0005/113756/minor.jpg) Wladek Minor
Wladek Minor![[M. Wall]](https://www.iucr.org/__data/assets/image/0019/113491/IMG_1078.jpg)
![[N. Johnson]](https://www.iucr.org/__data/assets/image/0017/113507/natalie_pic.jpg)
![[A. Foerster]](https://www.iucr.org/__data/assets/image/0017/113543/andreas_foerster.png)
![[H.J. Bernstein]](https://www.iucr.org/__data/assets/image/0020/113492/hjbernstein.jpg)
![[A. Goetz]](https://www.iucr.org/__data/assets/image/0018/113751/andy_gotz.jpg) Andrew Götz
Andrew Götz![[T.C. Terwilliger]](https://www.iucr.org/__data/assets/image/0015/113721/terwilliger.jpg)
![[S.J. Coles]](https://www.iucr.org/__data/assets/image/0005/113549/Simon_Coles.JPG)
![[S. Ward]](https://www.iucr.org/__data/assets/image/0020/113555/ward.jpg)
![[B. Matthews]](https://www.iucr.org/__data/assets/image/0010/113797/Matthews-Brian-09EC1353.jpg)
![[P. Aller]](https://www.iucr.org/__data/assets/image/0007/113875/IYDdlsMXPierreAller02.JPG)
![[B. McMahon]](https://www.iucr.org/__data/assets/image/0004/113737/bm.jpg)
![[G. Leonard]](https://www.iucr.org/__data/assets/image/0016/113506/GLeonard.jpg)
![[M. Blakely]](https://www.iucr.org/__data/assets/image/0005/113765/blakely.jpg)
![[K. Dziubek]](https://www.iucr.org/__data/assets/image/0015/113505/dziubek.jpg)
![[J.R. Hester]](https://www.iucr.org/__data/assets/image/0004/113692/James_Hester.jpg) James R. Hester
James R. Hester